One of the more famous images in biology is known as "Photo 51," an image of DNA that chemist Rosalind Franklin and Raymond Gosling created in 1952 by shooting X-rays through fibers of DNA and analyzing the patterns they left behind on film.
X-ray diffraction image of the double helix structure of the DNA molecule, taken in 1952. The image, famously known as "Photo 51," led to greater understanding of DNA and gave rise to the field of molecular biology. Image by Raymond Gosling/King's College London
One of the more famous images in biology is known as "Photo 51," an image of DNA that chemist Rosalind Franklin and Raymond Gosling created in 1952 by shooting X-rays through fibers of DNA and analyzing the patterns they left behind on film.
X-ray diffraction image of the double helix structure of the DNA molecule, taken in 1952. The image, famously known as "Photo 51," led to greater understanding of DNA and gave rise to the field of molecular biology. Image by Raymond Gosling/King's College London
That image provided Nobel Prize winners James Watson and Francis Crick with crucial information they needed to determine the 3D structure of DNA – knowledge that ultimately gave rise to molecular biology.
For much of the 20th century, this method of analyzing X-ray diffraction patterns has been the standard for determining the 3D structures of proteins, generally using a technique known as X-ray crystallography. But this method relies on getting proteins to pack tightly together to form uniform crystals, which is notoriously difficult, especially when it comes to the floppy, dynamic proteins that live in cell membranes.
Yet biologists are particularly interested in these membrane proteins because the membrane is the cell’s dock, its security checkpoint, its mailbox. From sugars to viral invaders to nerve impulses, all cellular traffic passes through the cell membrane, and likely interacts with the proteins embedded there, such as neurotransmitter and hormone receptors.
To understand all that transpires at the membrane, biologists need to zoom in to the atomic level. The proteins’ resistance to crystallization, therefore, left scientists in a bind.
Now, after decades of challenges, scientists have developed an imaging technique that allows them to visualize these tiny membrane proteins thousands of times smaller than the width of a human hair – and to do so without first having to form crystals.
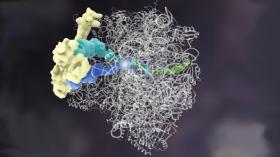
Complex, floppy molecules like this ribosome quality control complex were hard to see at the atomic level because their structures fell apart when researchers tried to prepare the sample for other visualizing techniques. UCSF researcher Adam Frost, PhD, was able to visualize it at last using a technique. Artistic rendering by Janet Iwasa, PhD UCSF.
Read more at University of California San Fransciso.