Inside a living cell, many important messages are communicated via interactions between proteins.
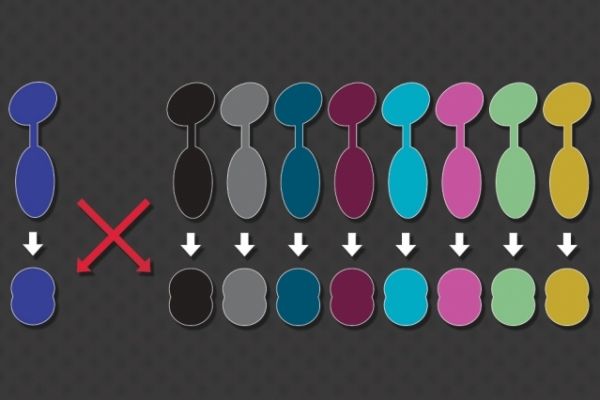
Inside a living cell, many important messages are communicated via interactions between proteins. For these signals to be accurately relayed, each protein must interact only with its specific partner, avoiding unwanted crosstalk with any similar proteins.
A new MIT study sheds light on how cells are able to prevent crosstalk between these proteins, and also shows that there remains a huge number of possible protein interactions that cells have not used for signaling. This means that synthetic biologists could generate new pairs of proteins that can act as artificial circuits for applications such as diagnosing disease, without interfering with cells’ existing signaling pathways.
“Using our high-throughput approach, you can generate many orthogonal versions of a particular interaction, allowing you to see how many different insulated versions of that protein complex can be built,” says Conor McClune, an MIT graduate student and the lead author of the study.
In the new paper, which appears today in Nature, the researchers produced novel pairs of signaling proteins and demonstrated how they can be used to link new signals to new outputs by engineering E. coli cells that produce yellow fluorescence after encountering a specific plant hormone.
Read more at Massachusetts Institute of Technology
Image: MIT researchers have found a way to generate many pairs of signaling proteins that don’t cross-talk with each other or with naturally occurring signaling proteins found in bacterial cells.
Image courtesy of the researchers