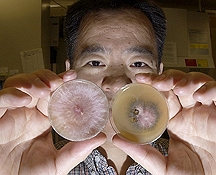
Purdue, Indiana - Why a pathogen is a pathogen may be answered as scientists study the recently mapped genetic makeup of a fungus that spawns the worst cereal grains disease known and also can produce toxins potentially fatal to people and livestock.
The fungus, which is especially destructive to wheat and barley, has resulted in an estimated $10 billion in damage to U.S. crops over the past 10 years. The scientists who sequenced the fungus' genes said that the genome will help them discover what makes this particular pathogen so harmful, what triggers the process that spreads the fungus and why various fungi attack specific plants.
Purdue, Indiana - Why a pathogen is a pathogen may be answered as scientists study the recently mapped genetic makeup of a fungus that spawns the worst cereal grains disease known and also can produce toxins potentially fatal to people and livestock.
The fungus, which is especially destructive to wheat and barley, has resulted in an estimated $10 billion in damage to U.S. crops over the past 10 years. The scientists who sequenced the fungus' genes said that the genome will help them discover what makes this particular pathogen so harmful, what triggers the process that spreads the fungus and why various fungi attack specific plants.
These investigations also may lead to producing plants that are completely resistant to the fungus Fusarium graminearum, something that hasn't been possible previously, said Jin-Rong Xu, a Purdue University molecular biologist. He is pinpointing which genes enable the fungus to cause the disease Fusarium head blight, or scab.
In a recent issue of the journal Science, Xu and an international scientific team reported that certain chromosomal regions in Fusarium graminearum appear to dictate plant and fungus molecular interactions that allow the fungus to contaminate crops and cause disease.
!ADVERTISEMENT!
The researchers located all of the genes on the fungus' chromosomes and then determined the genes' chemical makeup, or sequence.
"The Fusarium graminearum genome was easy to assemble because, unlike other fungal genomes, there aren't too many repetitive DNA sequences," Xu said. "It seems that this Fusarium can efficiently detect and remove duplicated sequences or transposable elements, which kept the genome clean and well-organized."
This basic information on the Fusarium graminearum genome will aid in further research and also provide information on other fungi and their interaction with plants, he said.
"Because we now have the genome sequence and a microarray containing the whole genome, it will help us determine what genes allow this fungus to behave as it does," Xu said. "It also will make it easier to identify and determine the function of similar genes in other pathogens and their plant interactions."
Fusarium graminearum, which exists worldwide, cuts crop yield, damages grain quality and produces mycotoxins. The fungus caused a widespread head blight epidemic during the 1990s in wheat- and barley-growing regions around the world. Experts estimate that from 1998 to 2000 the central and northern Great Plains of the United States suffered economic losses of $2.7 billion due to the disease. In Indiana alone in 1996, the fungus caused at least $38 million in crop loss, according to the USDA.
The mycotoxins caused by the fungus can affect people and livestock that ingest infected grain. Pigs, cattle, horses, poultry and people can develop vomiting, loss of appetite, diarrhea, staggering, skin irritation and immunosuppression. The most severe cases can be fatal.
Some scientific evidence suggests that these toxins cause cancer. People in developing countries are at the greatest risk of eating grain contaminated with Fusarium mycotoxins. Although not all types of Fusarium cause disease and produce toxins, those types that do infect other crops, including corn and hay.
Currently, fungicides aren't effective because the fungus only attacks during the beginning of the plants' flowering stage. It's difficult to gauge the precise time to spray, and it's expensive to try to protect the crops over a long period. The fungus can survive through the winter in crop remnants left in fields as natural mulch.
The pathogen is most likely to appear and cause infection in early spring when the weather is warm and humid or rainy. By the time Fusarium contamination is noticeable on plants, head blight has already damaged the grain.
Xu is searching for the genes involved in the infection process.
"We are using the whole-genome microarray of Fusarium graminearum to identify the genes that are functional during plant infection," Xu said. "We are looking at the biochemical signaling pathways that influence whether a gene is turned on or off. This will help us find ways to develop new, stable and environmentally safe ways to prevent these infections."
Xu was one of the co-applicants for a $1.9 million grant from a U.S. Department of Agriculture/National Science Foundation partnership that funded the genome project. The endeavor was headed by Corby Kistler, a USDA-Agricultural Research Service geneticist based at the University of Minnesota.
Christina Cuomo of the Broad Institute at the Massachusetts Institute of Technology led the sequencing. Other members of the research team included scientists from Michigan State University; Cornell University; Pacific Northwest National Laboratory; University of Arizona; St. Louis University; University of Tennessee; and institutions in Germany, Canada, Austria, England, France, Ukraine and the Netherlands.
Writer: Susan A. Steeves, (765) 496-7481,